iGenomics app for iPhone and iPad
Developer: Michael Schatz
First release : 17 Mar 2015
App size: 5.71 Mb
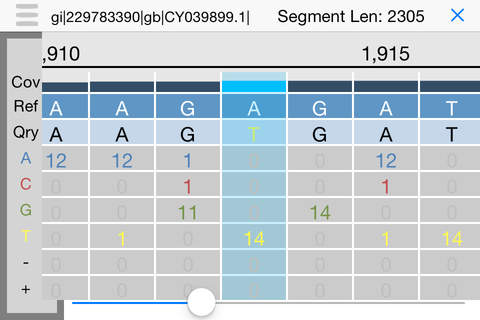
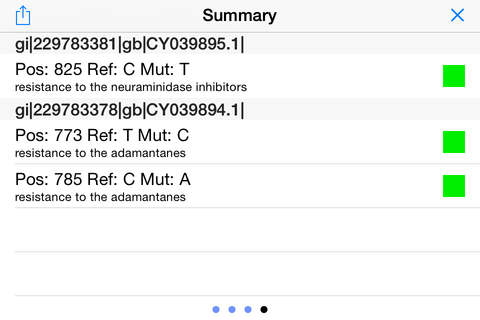
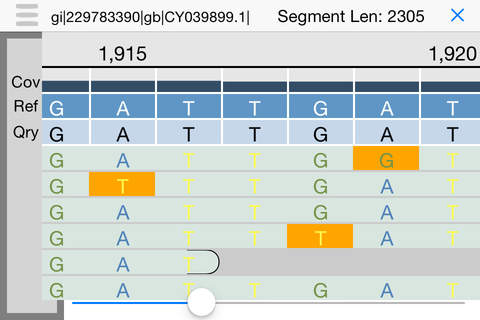
iGenomics is a scientific application for studying and analyzing DNA sequences. It allows you to map DNA sequencing reads from a sample to a reference genome to determine mutations, such as matching the DNA sequencing reads from a virus or microbe to a known reference. After aligning the reads, iGenomics displays the results in an interactive view, showing how each read aligns along with a coverage profile that highlights any detected mutations. It also can load and display lists of known mutations to quickly screen samples for their most important positions. iGenomics follows the state-of-the-art algorithms for alignment and genotyping, and has been validated with Illumina, Ion Torrent, PacBio, and Oxford Nanopore sequencing data with standard pipelines including Bowtie, BWA, and SAMtools.
Please visit http://schatzlab.cshl.edu/iGenomics/ for more information.